Demo example - finger movements#
The corresponding jupyter notebook script can be downloaded here.
The necessary input data (surfaces + muscles description) can be downloaded here:.
Import libraries#
We start by importing necessary libraries. Myoelectric digital twin is
part of the neurodec software. Apart from standard auxiliary
libraries we also need meshio library to work with .stl surfaces
%config Completer.use_jedi = False
import glob
import os
import json
import matplotlib.pyplot as plt
import trimesh
import numpy as np
import neurodec as nd
nd.mdt._API_URL = "http://35.173.188.106:8001/"
nd.mdt.wait = False # Set to true to wait step by step.
Load model surfaces#
Subject’s forearm anatomy is represented by .stl meshes of the
different tissue surfaces: upper skin, lower skin, individual muscles
and bones.
The surface filename for each muscle is stored in
muscles_description.json file.
We load each mesh with meshio package and create a nd.mdt.Surface
for each tissue surface.
def load_surface(filename: str):
mesh = trimesh.load_mesh(filename)
return mesh.vertices, mesh.faces
MODEL_DIR = "<your-data-path>/right-arm-template"
muscles_description_fname = os.path.join(MODEL_DIR, 'muscles_description.json')
with open(muscles_description_fname) as f:
muscles_description = json.load(f)
bones = []
for filename in glob.glob(os.path.join(MODEL_DIR, 'bones', '*.stl')):
vertices, triangles = load_surface(filename)
bones.append(nd.mdt.Surface.new(vertices, triangles, nd.mdt.SurfaceType.BONE))
muscles = []
for muscle in muscles_description['muscles']:
fname = os.path.join(MODEL_DIR, 'muscles', muscle['fname'])
vertices, triangles = load_surface(fname)
muscles.append(nd.mdt.Surface.new(vertices, triangles, nd.mdt.SurfaceType.MUSCLE, label=muscle['label']))
skins = []
filename = os.path.join(MODEL_DIR, 'skin', 'inner.stl')
vertices, triangles = load_surface(filename)
skins.append(nd.mdt.Surface.new(vertices, triangles, nd.mdt.SurfaceType.INNER_SKIN))
filename = os.path.join(MODEL_DIR, 'skin', 'outer.stl')
vertices, triangles = load_surface(filename)
skins.append(nd.mdt.Surface.new(vertices, triangles, nd.mdt.SurfaceType.OUTER_SKIN))
Generate volume conductor#
Volume conductor is a function of anatomy surfaces and conductivities per tissue. Here, the default conductivity values are used.
conductor = nd.mdt.Conductor.new(bones + muscles + skins)
Generate bracelet electrodes#
We generate a bracelet of electrodes made of ten rings. Each ring has 16 equidistant electrodes of 3mm radius. Distance between rings is 9 mm.
Note that user can create electrodes by directly providing their centers
and radii with nd.mdt.Electrode.new(location, radius).
outer_skin_surface = skins[1]
radius = 0.003 # in meters
first_electrode_location = np.array([-0.247705, -0.083379, 0.838751])
rings_normal = np.array([0.17797643704463834, 0.19429176558661732, 0.9646632042750106])
n_rings = 10
distance_between_rings = 0.009 # in meters
n_electrodes_per_ring = 16
bracelet = nd.mdt.ElectrodeBracelet.new(
outer_skin_surface, radius, first_electrode_location, rings_normal,
n_rings, distance_between_rings, n_electrodes_per_ring)
You can save generated electrodes as spheres with trimesh for futher
visualization
print(bracelet.electrodes[0].location)
for i, electrode in enumerate(bracelet.electrodes):
sphere = trimesh.creation.icosphere()
sphere.vertices = sphere.vertices * electrode.radius
sphere.vertices = sphere.vertices + electrode.location
sphere.export(MODEL_DIR + f'/elec_{i}.stl')
Generate forward solver#
Forward solver is a function of volume conductor and electrodes
forward_solution = nd.mdt.ForwardSolution.new(conductor, bracelet.electrodes)
Generate fiber geometry#
User can directly provide the number of fibers, or it can be automatically estimated from the average fiber radius.
Start and ending planes for fibers are stored in the
muscle_description.
fibers_per_muscle = []
for i, muscle in enumerate(muscles):
plane_origins = muscles_description['muscles'][i]['plane_origins']
plane_normals = muscles_description['muscles'][i]['plane_normals']
fibers = nd.mdt.Fibers.new(muscle, plane_origins, plane_normals)
fibers_per_muscle.append(fibers)
We also add individual finger “muscle” surfaces. Note, that those are not anatomical muscles but a subdivision of muscles controlling fingers so that individual finger motor units could be generated.
finger_surfaces_description_fname = os.path.join(MODEL_DIR, 'individual_finger_surfaces.json')
with open(finger_surfaces_description_fname) as f:
finger_surfaces_description = json.load(f)
for muscle in finger_surfaces_description['muscles']:
# load extra surfaces for individual fingers
fname = os.path.join(MODEL_DIR, 'muscles', muscle['fname'])
vertices, triangles = load_surface(fname)
muscles.append(nd.mdt.Surface.new(vertices, triangles, nd.mdt.SurfaceType.MUSCLE, label=muscle['label']))
# Generate corresponding fibers
plane_origins = muscle['plane_origins']
plane_normals = muscle['plane_normals']
fibers = nd.mdt.Fibers.new(muscles[-1], plane_origins, plane_normals)
fibers_per_muscle.append(fibers)
Generate fiber basis#
Fiber basis is a function of fibers and forward solution.
fiber_basis_per_muscle = []
for i, fibers in enumerate(fibers_per_muscle):
fiber_basis = nd.mdt.FiberBasis.new(fibers, forward_solution)
fiber_basis_per_muscle.append(fiber_basis)
Compute MUAPs#
To compute MUAPs for each muscle, fiber properties and motor units (MUs) should be generated and a sampling frequency (in Hz) should be provided.
Here, fiber properties are generated using default tendon, velocity and neuromascular junction values.
The number of MU should be specified. Here, default values for minimum and maximum relative MU areas are used.
SAMPLING_FREQUENCY = 8000
n_motor_units = 150
muaps_per_muscle = []
for fibers, fiber_basis in zip(fibers_per_muscle, fiber_basis_per_muscle):
fiber_properties = nd.mdt.FiberProperties.new(fibers)
motor_units = nd.mdt.MotorUnits.new(fibers, n_motor_units)
muaps = nd.mdt.MotorUnitsActionPotentials.new(fibers, fiber_basis, fiber_properties, motor_units,
SAMPLING_FREQUENCY)
muaps_per_muscle.append(muaps)
Simulate finger movements#
Print the list of muscle labels
for i, muscle in enumerate(muscles):
print(i, muscle.label)
0 Extensor carpi radialis longus
1 Extensor carpi radialis brevis
2 Extensor carpi ulnaris
3 Flexor carpi radialis
4 Palmaris longus
5 Flexor carpi ulnaris humeral head
6 Flexor carpi ulnaris ulnar head
7 Pronator teres humeral head
8 Pronator teres ulnar head
9 Pronator quadratus
10 Supinator
11 Brachioradialis
12 Flexor digitorum superficialis humeroulnar head
13 Flexor digitorum superficialis radial head
14 Abductor pollicis longus
15 Anconeus
16 Extensor digiti minimi
17 Extensor digitorum
18 Extensor indicis
19 Extensor pollicis brevis
20 Extensor pollicis longus
21 Flexor digitorum profondus
22 Flexor pollicis longus
23 Extensor digitorum INDEX
24 Extensor digitorum MIDDLE
25 Extensor digitorum RING
26 Flexor digitorum profondus INDEX
27 Flexor digitorum profondus MIDDLE
28 Flexor digitorum profondus RING
29 Flexor digitorum profondus PINKY
30 Flexor digitorum superficialis INDEX
31 Flexor digitorum superficialis MIDDLE
32 Flexor digitorum superficialis RING
33 Flexor digitorum superficialis PINKY
Select muscle indices for desired movements. Here are some examples:
movements = {
# Thumb flexion: FPL
'thumb_flex': [22],
# Thumb abduction: APL, EPB
'thumb_abduct': [14, 19],
# Thumb extension (lift): EPL
'thumb_ext': [20],
# Index flexion: FDP index, FDS index
'index_flex': [26, 30],
# Index extension: EI, EDC index
'index_ext': [18, 23],
# Middle flexion: FDP middle, FDS middle
'middle_flex': [27, 31],
# Middle extension: EDC middle
'middle_ext': [24],
# Ring flexion: FDP ring, FDS ring
'ring_flex': [28, 32],
# Ring extension: EDC ring
'ring_ext': [25],
# Pinky flexion: FDP pinky, FDS pinky
'pinky_flex': [29, 33],
# Pinky extension: EDM
'pinky_ext': [16]
}
Check the target muscles
for movement, target_muscles in zip(movements.keys(), movements.values()):
print(movement)
for muscle_id in target_muscles:
print('\t' + muscles[muscle_id].label)
thumb_flex
Flexor pollicis longus
thumb_abduct
Abductor pollicis longus
Extensor pollicis brevis
thumb_ext
Extensor pollicis longus
index_flex
Flexor digitorum profondus INDEX
Flexor digitorum superficialis INDEX
index_ext
Extensor indicis
Extensor digitorum INDEX
middle_flex
Flexor digitorum profondus MIDDLE
Flexor digitorum superficialis MIDDLE
middle_ext
Extensor digitorum MIDDLE
ring_flex
Flexor digitorum profondus RING
Flexor digitorum superficialis RING
ring_ext
Extensor digitorum RING
pinky_flex
Flexor digitorum profondus PINKY
Flexor digitorum superficialis PINKY
pinky_ext
Extensor digiti minimi
Let’s define higher level function to simulate specific muscle activation patterns
def assemble_emg(emg_list):
max_length = np.max([e.shape[1] for e in emg_list])
emg_total = np.zeros([emg_list[0].shape[0], max_length])
for emg in emg_list:
n_samples = emg.shape[1]
emg_total[:, :n_samples] += emg
return emg_total
def single_muscle_activation(max_mvc, duration, peak_time, fs):
'''Generates a single activation pattern smoothly rising to the max MVC and back to zero'''
n_time_samples = int((peak_time + duration / 2.) * fs)
t = np.linspace(-np.pi / 2, np.pi + np.pi / 2, int(duration * fs))
waveform = max_mvc * 0.5 * (np.sin(t) + 1.)
i_0 = n_time_samples - len(waveform)
muscle_activation = np.zeros(n_time_samples)
muscle_activation[i_0:] = waveform
return muscle_activation
def generate_muscle_activations(n_muscles, mean_max_mvc, std_max_mvc, duration, peak_time, fs):
'''Generates a multilple activation patterns with some variability in max MVC per muscle'''
muscle_activations = []
for i in range(n_muscles):
max_mvc = np.random.uniform(mean_max_mvc - std_max_mvc,
mean_max_mvc + std_max_mvc)
max_mvc = np.clip(max_mvc, 0., 100.)
muscle_activations.append(single_muscle_activation(max_mvc, duration, peak_time, fs))
return muscle_activations
def generate_movement(target_muscles, muaps_per_muscle,
mean_max_mvc, std_max_mvc,
duration, peak_time, fs, random_seed=42):
'''Generates EMG corresponding to a specific movement'''
# Generate muscle activations
n_muscles = len(target_muscles)
muscle_activations = generate_muscle_activations(n_muscles, mean_max_mvc, std_max_mvc,
duration, peak_time, fs)
# Generate impulse trains
impulse_trains_per_muscle = []
for muscle_id, single_activation in zip(target_muscles, muscle_activations):
impulse_trains_per_muscle.append(nd.mdt.ImpulseTrains.new(muaps_per_muscle[muscle_id],
single_activation,
random_seed=random_seed))
# Generate EMG for each muscle
emg_per_muscle = []
for impulse_trains in impulse_trains_per_muscle:
emg = nd.mdt.Electromyography.new(impulse_trains)
emg.wait() # wait for the computation to finish before loading the EMG data
emg_per_muscle.append(emg.data.copy())
# Sumup EMG signals of induvidual muscles
emg_total = assemble_emg(emg_per_muscle)
return muscle_activations, emg_total
Example: 2 seconds long Index extension with 40%MVC peaking at 1.5 sec#
target_muscles = movements['index_ext']
mean_max_mvc = 40 # %MVC
std_max_mvc = 10 # add some inter muscle variability in %MVC
duration = 2. # sec
peak_time = 1.5 # sec
muscle_activations, emg = generate_movement(target_muscles, muaps_per_muscle,
mean_max_mvc, std_max_mvc,
duration, peak_time, SAMPLING_FREQUENCY,
random_seed=42)
# plot muscle activations
plt.figure()
for single_activation in muscle_activations:
times = np.arange(len(single_activation)) / SAMPLING_FREQUENCY
plt.plot(times, single_activation)
# plot EMG
times = np.arange(emg.shape[1]) / SAMPLING_FREQUENCY
plot_shift = np.ptp(emg)
rings_to_plot = [0, 4, 9]
for ring in rings_to_plot:
emg_to_plot = emg[ring*16:(ring+1)*16]
plt.figure()
plt.plot(times, emg_to_plot.T + plot_shift * np.arange(16))
plt.show()
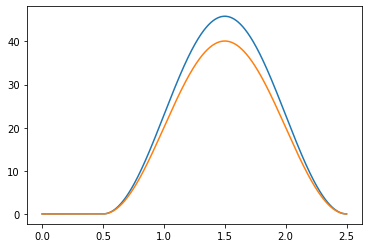
Muscle activations#
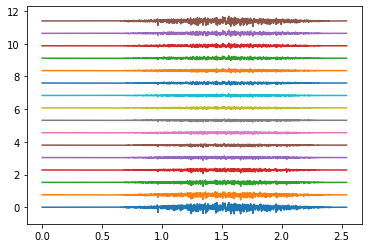
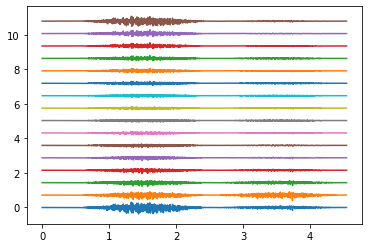
Electrodes ring 0#
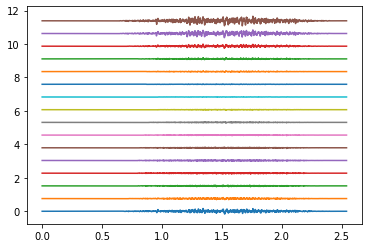
Electrodes ring 4#
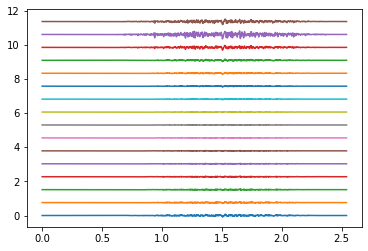
Electrodes ring 9#
Example: Index extension followed by pinky extension#
# Index
target_muscles = movements['index_ext']
mean_max_mvc = 40 # %MVC
std_max_mvc = 10 # add some inter muscle variability in %MVC
duration = 2. # sec
peak_time = 1.5 # sec
_, emg_index = generate_movement(target_muscles, muaps_per_muscle,
mean_max_mvc, std_max_mvc,
duration, peak_time, SAMPLING_FREQUENCY,
random_seed=42)
# Pinky
target_muscles = movements['pinky_ext']
mean_max_mvc = 40 # %MVC
std_max_mvc = 10 # add some inter muscle variability in %MVC
duration = 2. # sec
peak_time = 3.5 # sec
_, emg_pinky = generate_movement(target_muscles, muaps_per_muscle,
mean_max_mvc, std_max_mvc,
duration, peak_time, SAMPLING_FREQUENCY,
random_seed=42)
# Total
emg_total = assemble_emg([emg_index, emg_pinky])
# plot EMG
times = np.arange(emg_total.shape[1]) / SAMPLING_FREQUENCY
plot_shift = np.ptp(emg_total)
rings_to_plot = [0, 4, 9]
for ring in rings_to_plot:
emg_to_plot = emg_total[ring*16:(ring+1)*16]
plt.figure()
plt.plot(times, emg_to_plot.T + plot_shift * np.arange(16))
plt.show()
Electrodes ring 0#
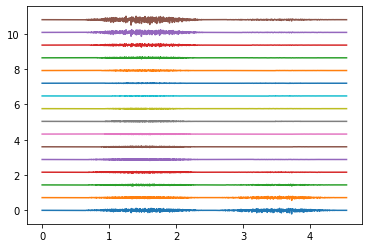
Electrodes ring 4#
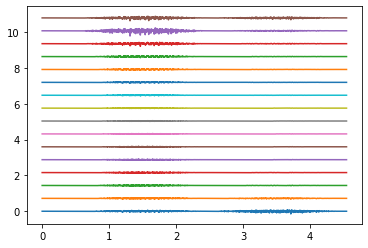
Electrodes ring 9#
Example: Simultaneous index and thumb flexion#
# Index
target_muscles = movements['index_flex']
mean_max_mvc = 40 # %MVC
std_max_mvc = 10 # add some inter muscle variability in %MVC
duration = 2. # sec
peak_time = 1.5 # sec
_, emg_index = generate_movement(target_muscles, muaps_per_muscle,
mean_max_mvc, std_max_mvc,
duration, peak_time, SAMPLING_FREQUENCY,
random_seed=42)
# Thumb
target_muscles = movements['thumb_flex']
mean_max_mvc = 40 # %MVC
std_max_mvc = 10 # add some inter muscle variability in %MVC
duration = 2. # sec
peak_time = 1.5 # sec
_, emg_thumb = generate_movement(target_muscles, muaps_per_muscle,
mean_max_mvc, std_max_mvc,
duration, peak_time, SAMPLING_FREQUENCY,
random_seed=42)
# Total
emg_total = assemble_emg([emg_index, emg_thumb])
# plot EMG
times = np.arange(emg_total.shape[1]) / SAMPLING_FREQUENCY
plot_shift = np.ptp(emg_total)
rings_to_plot = [0, 4, 9]
for ring in rings_to_plot:
emg_to_plot = emg_total[ring*16:(ring+1)*16]
plt.figure()
plt.plot(times, emg_to_plot.T + plot_shift * np.arange(16))
plt.show()
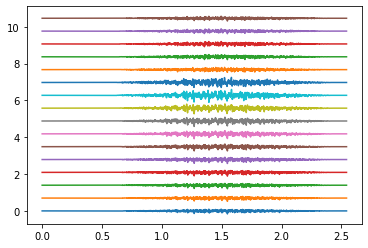
Electrodes ring 0#
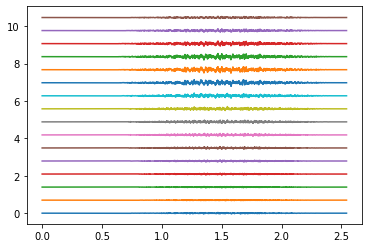
Electrodes ring 4#
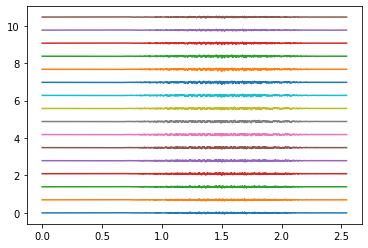
Electrodes ring 9#